Epigenome Mapping Reveals Distinct Modes of Gene Regulation and Widespread Enhancer Reprogramming by the Oncogenic Fusion Protein EWS-FLI1 - ScienceDirect

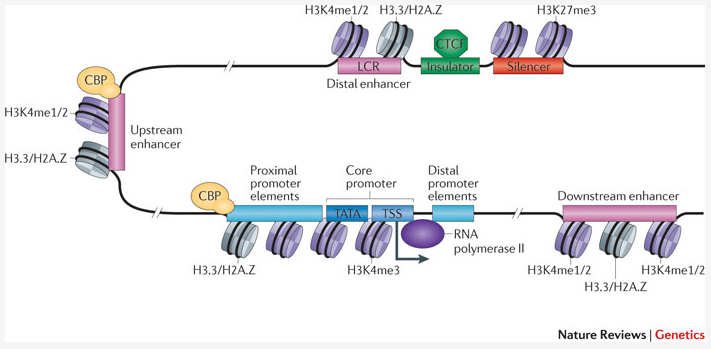
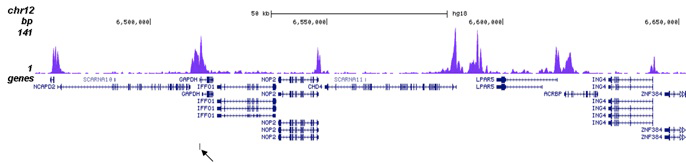
Poised and active enhancers. (A) H3K4me1 and H3K27Ac ChIP-seq tags from... | Download Scientific Diagram
Atheroprotective Flow Upregulates ITPR3 (Inositol 1,4,5-Trisphosphate Receptor 3) in Vascular Endothelium via KLF4 (Krüppel-L
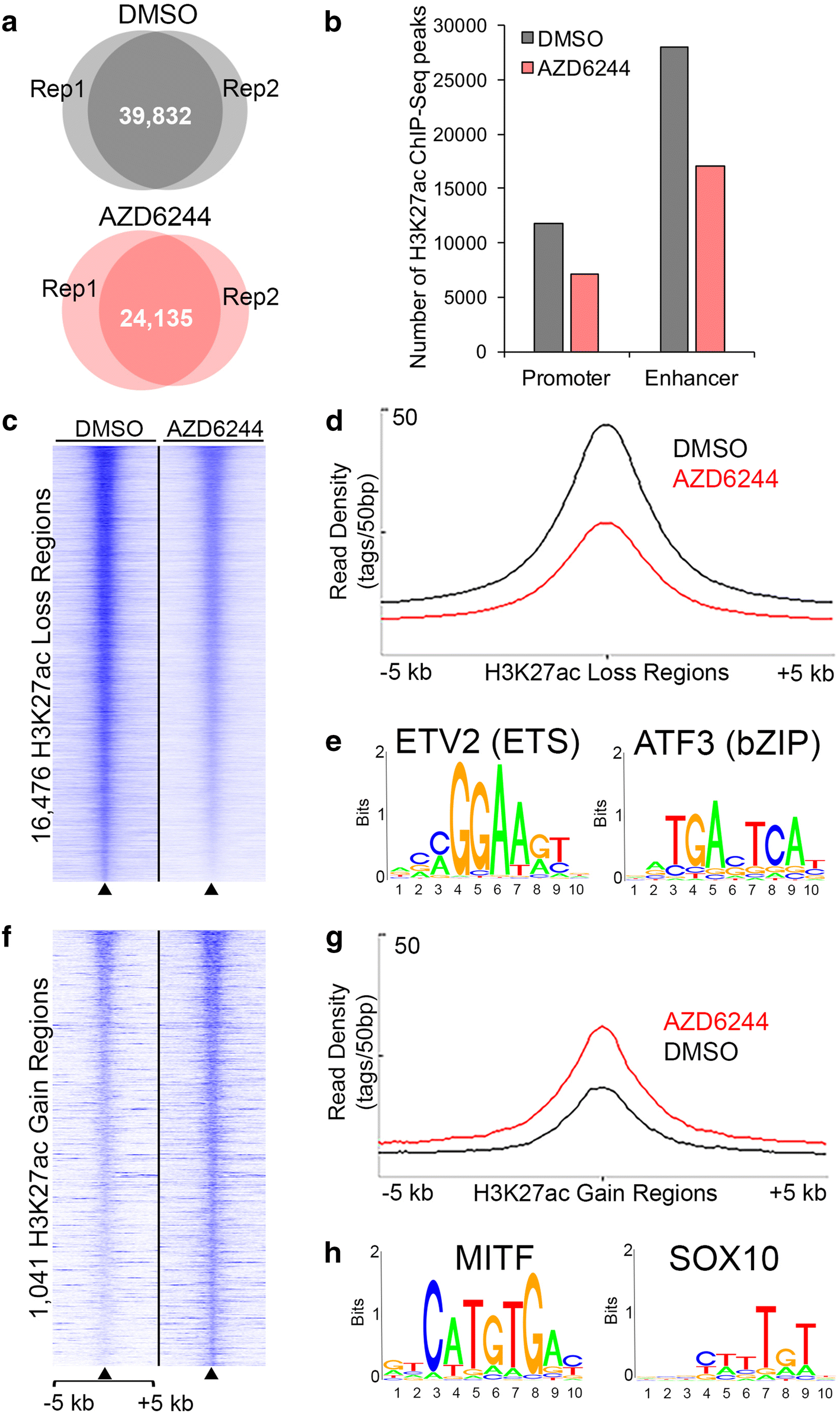
MEK inhibition remodels the active chromatin landscape and induces SOX10 genomic recruitment in BRAF(V600E) mutant melanoma cells | Epigenetics & Chromatin | Full Text

Genome-Wide Mapping of DNA Accessibility and Binding Sites for CREB and C/EBPβ in Vasopressin-Sensitive Collecting Duct Cells | American Society of Nephrology
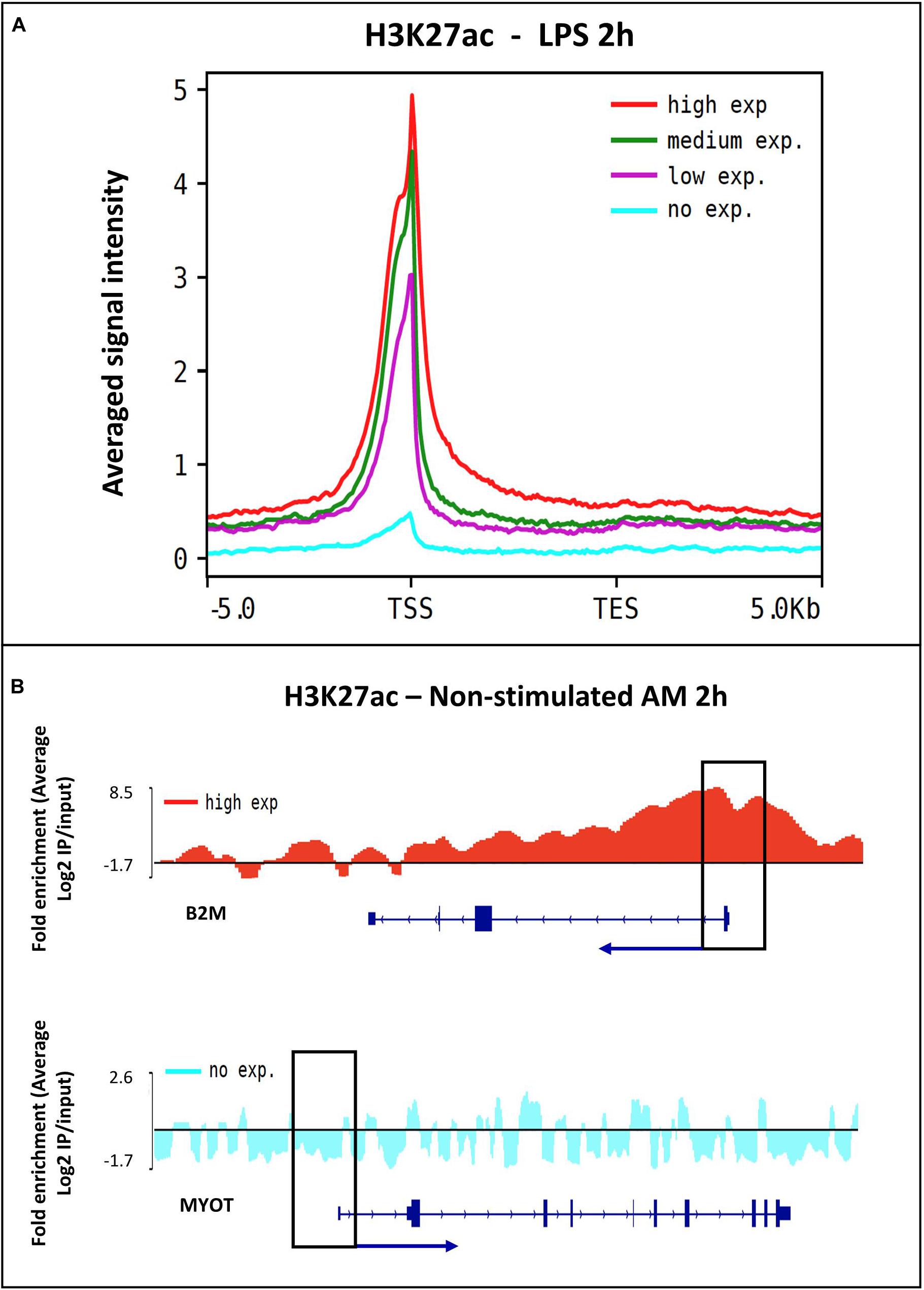
Frontiers | Changes in H3K27ac at Gene Regulatory Regions in Porcine Alveolar Macrophages Following LPS or PolyIC Exposure

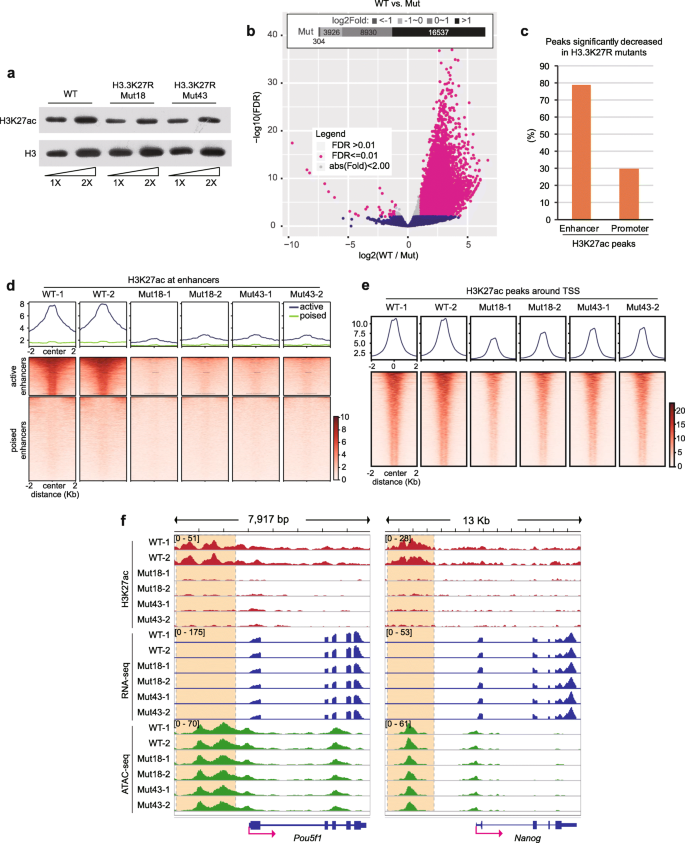
Phosphorylation of histone H3.3 at serine 31 promotes p300 activity and enhancer acetylation. - Abstract - Europe PMC

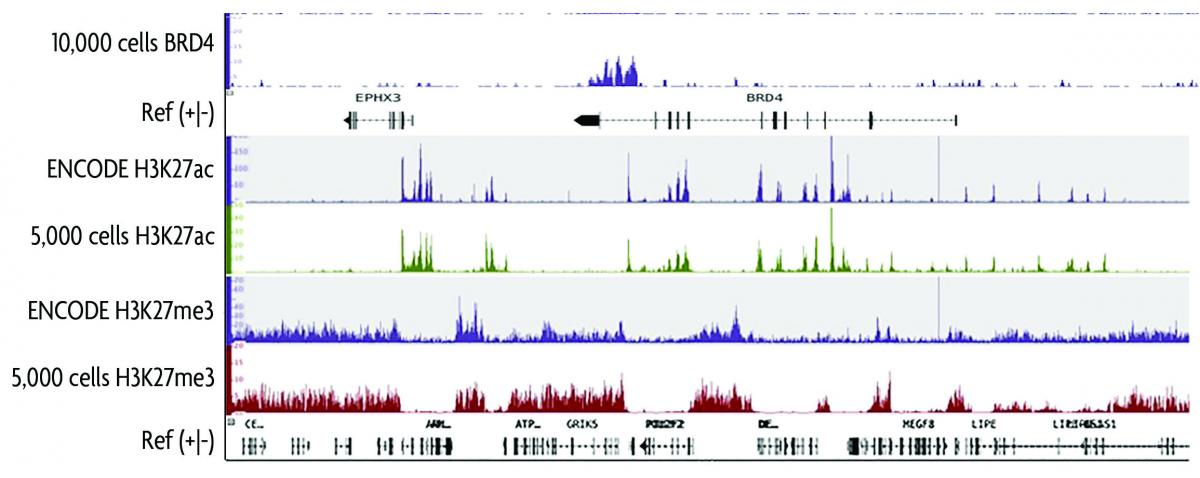
FiTAc-seq: fixed-tissue ChIP-seq for H3K27ac profiling and super-enhancer analysis of FFPE tissues | Nature Protocols

Histone H3K4me1 and H3K27ac play roles in nucleosome depletion and eRNA transcription, respectively, at enhancers | bioRxiv
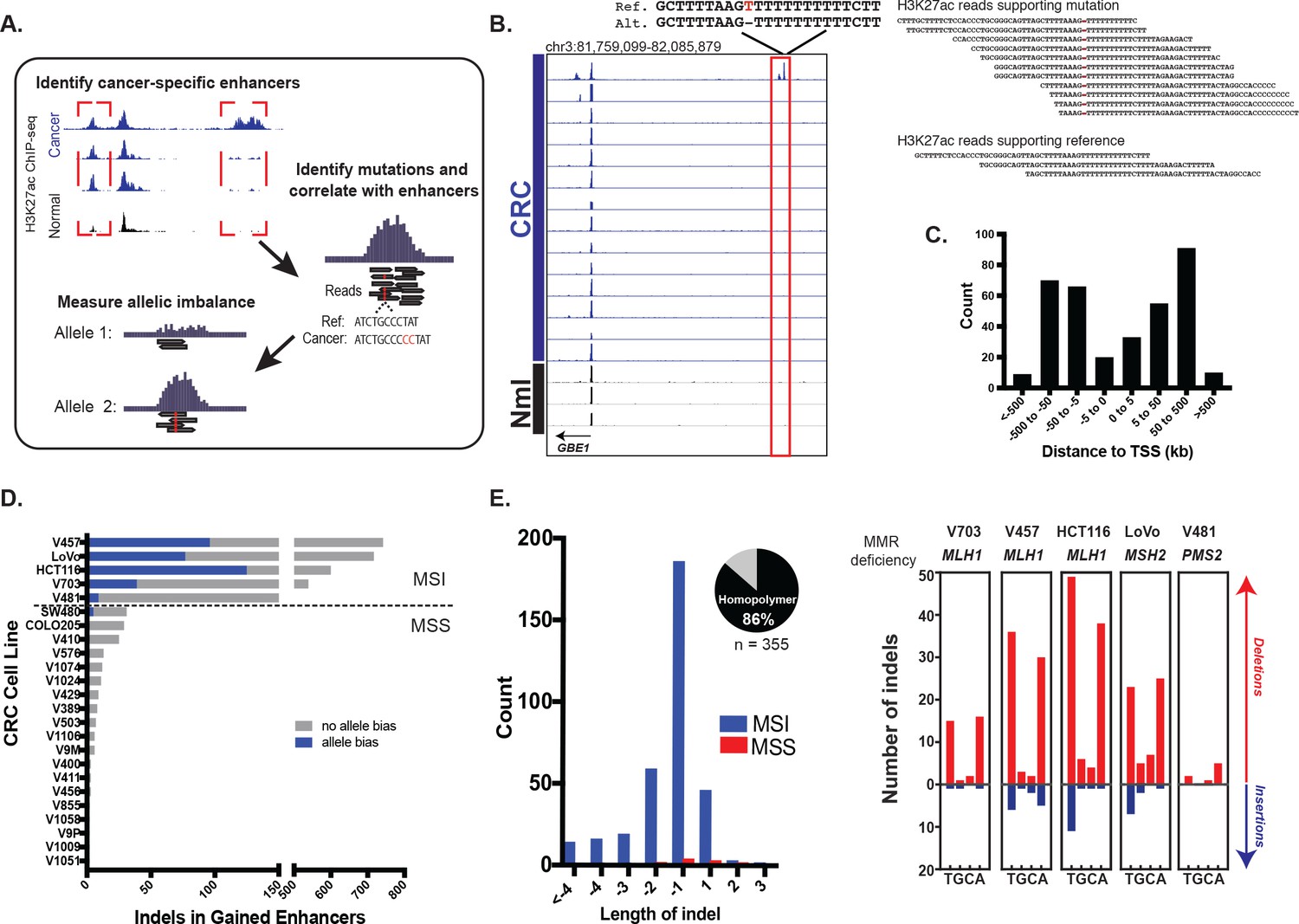
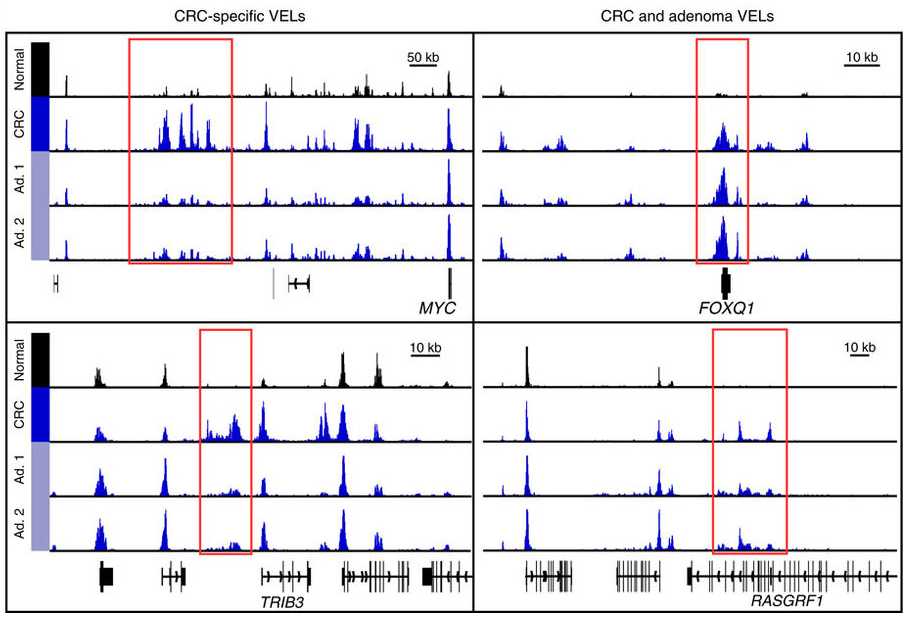
Figures and data in Mismatch repair-signature mutations activate gene enhancers across human colorectal cancer epigenomes | eLife
Genome-Wide Analysis of Histone Modifications: H3K4me2, H3K4me3, H3K9ac, and H3K27ac in Oryza sativa L. Japonica

Integrated Analysis of Whole-Genome ChIP-Seq and RNA-Seq Data of Primary Head and Neck Tumor Samples Associates HPV Integration Sites with Open Chromatin Marks. - Abstract - Europe PMC